
일 시
| 2013년 8월 26일(월)- 30일(금)
장 소 | 서울의대 동창회관 3층 가천홀(월~목), 서울의대 의학도서관 3층 전산실습실(금, 8/30)
주 관
|
서울의대 정보의학실, 시스템 바이오 정보의학 연구센터 (SBI-NCRC)
주 최 |
대한의료정보학회, 한국생물정보시스템생물학회
제5차 Genome Data Analysis
Workshop을 개최하며
|
안녕하십니까?
Genome Data Analysis 웍샵이 그 다섯 번 째에 접어듭니다. 우리도 단순한 강좌 시리즈 보다 실제적으로 실행가능한 코드를 중심으로 실습, finger exercise를 할 수 있는 프로그램을 만들어보자는 바램으로 시작한지도 3년에 되어갑니다. 그간 보내주신 성원에 깊이 감사드립니다.
Genome Data는 나날이 늘어갑니다. 1000 Genome Project가 그 하나고, TCGA (The Cancer Genome Atlas)가 다른하나입니다. 그 외에도 다양한 사업을 통해 Genome Data가 널리 공개되어, 이제는 "유전체 연구의 민주화 시대" 가 만개했습니다. 저는 학회 등에서 공공연히, 그리고 용감하게 "앞으로 10년 후면, 암이 만성질환화 될 것입니다"라고 이야기하곤 합니다. 허황된 이야기가 아니라고 생각합니다. 암은 근본적으로 DNA의 질환이라고 받아들여집니다. 눈 부신 기술발전으로 DNA의 비밀이 드러나고 있습니다. 다양한 진단-치료 전략이 발전하고 있고, 머지 않아 상당 부분의 암종은, 지금처럼 급성 질환이 아닌 만성질환의 하나로 자리매김할 것임을 믿어 의심치 않습니다.
많은 분들이 “맞춤의학”을 이야기 합니다. 맞춤의학에 대한 제 정의는 개개인 별로 다른 다양한 데이터에 기반해서 개인별로 특화된 의학적 전략을 적용하는 분야입니다. 바 로 임상 데이터와 유전체 데이터에 기반한 데이터 의학이지요. 그 어느 때보다도 Genomic Data에 대한 올바른 이해가 중요한 시대가 되었습니다. 연구자들의 실질적 문제해결에 도움이 되기 위해서, 서울의대 정보의학실과 서울대 시스템 바이오 정보의학 국가핵심연구센터에서는 2013년도 하계 방학을 맞아 초보자도 접근할 수 있는 실용적인 유전체 데이터 분석의 전반적인 기초지식을 연습하고, 연구 뿐 아니라 맞춤의료 및 산업에 응용가능한 내용으로 GDA (Genome Data Analysis) 웍샵을 개설했습니다. 본 웍샵을 통해 실용적인 유전체 정보 분석의 역량을 강화하는 기회가 되시기를 기대하며 많은 관심과 참여를 부탁드립니다.
2013년 6월, 서울대 시스템 바이오 정보의학 연구센터장 김 주 한
|
|
|
제1차 GDA Workshop: 2011년 8월 22일~26일, 서울의대
제2차 GDA Workshop: 2012년 2월 20일~24일, 서울의대
제2차 웍샵에서는 다음과 같은 새로운 실습모듈 3개가 추가 되었다.
(1) micro-RNA 데이터 분석
(2) 개인유전체 해석: Personal Genome Interpretation
(3) 암유전체/희귀질환유전체 데이터 분석
제3차 GDA Workshop: 2012년 8월 20일~24일, 서울의대
제 3차 웍샵에서는 다음과 같은 2개의 실습모듈이 추가되었다.
(1) Family-based 엑솜시퀀싱 분석
(2) TCGA (The Cancer Genome Atlas) 데이터 분석
제4차 GDA Workshop: 2013년 2월 18일~22일, 서울의대
4차 웍샵에서는 다음과 같은 2개의 실습모듈이 추가되었다.
(1) eQTL 데이터 분석
(2) PheWAS & EWAS 데이터 분석
본5차 웍샵에서는 다음과 같은 3개의 실습모듈이 추가될 예정이다.
(1)
시퀀스 레벨 전사체 분석: Isoforms, Alternative Splicing, RNA-editing, and Fusion Gene
(2)
개인유전체 해석을 위한 지식/데이터기반 자원 소개와 유전적 위험 예측 분석
(3)
Post-GWAS: EMR 데이터와 질병 연관 분석
|

실습서 "유전체 데이터 분석" 출간
|
|
강좌 일정
|
강좌일정은 주최측의 사정에 따라 변경될 수 있습니다.
|
DAY 1: Advanced Microarray Data Analysis
|
|
8월 26일(월)
|
|
시간
|
주 제
|
강 사
|
|
8:30 ~ 9:30
|
등록 및 사전 프로그램 설치 |
|
9:30 ~ 9:50
|
Advanced Microarray Data Analysis |
김주한 교수
|
|
9:50 ~ 10:40
|
Gene Expression Analysis
- Normalization
- Differential Expression Analysis
- Classification Analysis
|
김주한 교수
(서울의대)
|
|
10:50 ~ 12:10
|
실 습 I: Bioconductor
t-test, SAM, ANOVA, FDR
|
이수연s, 백수연
|
|
12:10 ~ 13:10
|
중 식
|
|
13:10 ~ 14:00
|
Classification and Clustering
- Classification Analysis
- Clustering Analysis
- Evaluation and Validation
|
손경아 교수
(아주대)
|
|
14:10 ~ 15:30
|
실 습 II: KNN, SOM, HC, PCA
LDA, DTs, SVMs
|
이수연, 서희원
|
|
15:40 ~ 16:30
|
Gene-set Approaches & Prognostic Subgroup Prediction
- Gene Ontology & Pathway Analysis
- Gene Set Enrichment Analysis
- Prognostic Subgroup Prediction
|
조성범 박사
(국립보건연구원)
|
|
16:40 ~ 18:00
|
실 습 III: Gene Set Enrichment Analysis
Cox-PH, Log Rank Test
David, ArrayXPath
|
김기태, 백수연
|
|
DAY 2: Next Generation Sequencing & Personal Genome Data Analysis
|
|
8월 27일(화)
|
|
시간
|
주 제
|
강 사
|
|
8:30 ~ 9:30
|
등록 및 사전 프로그램 설치 |
|
9:30 ~ 9:50
|
Next Generation Sequencing & Personal Genome Data Analysis
|
김주한 교수
|
|
9:50 ~ 10:40
|
NGS Platforms and Applications
- Current NGS Platforms
- NGS Data Formats
- NGS Data Analysis Technologies
- NGS Applications
|
김지훈 박사
(마크로젠)
|
|
10:50 ~ 12:10
|
실 습 I: NGS Data Processing
NGS Data Format Converting
NGS Visualization Tools
|
서희원, 임재현
|
|
12:10 ~ 13:10
|
중 식
|
|
13:10 ~ 14:00
|
NGS Data Analysis
- Sequence Alignment Algorithms
- Whole Genome and Exome Data Analysis
- Variation Detection and Reference Genome
|
이수연 박사
(서울의대)
|
|
14:10 ~ 15:30
|
실 습 II: Exome Sequencing Alignment
SNP and Indel Identification
Variation Filtering
|
박찬희, 서희원
|
|
15:40 ~ 16:30
|
Personal Genome Interpretation
- Phenotype Annotation
- Genetic Risk Prediction
- Healthcare Application
|
김주한 교수
(서울의대)
|
|
16:40 ~ 18:00
|
실 습 III: SNP Prioritization
Genetic Risk Prediction methods
Resources for Personal Genome Interpretation
(dbGAP, PheGeni, SNPedia, PhenoDB)
|
이수연s, 백수연
|
|
DAY 3: RNA-seq Data Analysis
|
|
8월 28일(수)
|
|
시간
|
주 제
|
강 사
|
|
8:30 ~ 9:30
|
등록 및 사전 프로그램 설치 |
|
9:30 ~ 9:50
|
RNA-seq Data Analysis
|
김주한 교수
|
|
9:50 ~ 10:40
|
RNA-Seq Expression Profile Analysis
- Read Alignment Methods
- Expression Quantification Strategy
- Differentially Expressed Genes Identification
- Expression Profile Analysis
|
정제균 박사
(삼성유전체연구소)
|
|
10:50 ~ 12:10
|
실 습 I: Read alignment with TopHat,
Expression Quantification with Cufflinks
RNA-Seq Gene Expression Analysis
|
임재현, 서희원
|
|
12:10 ~ 13:10
|
중 식
|
|
13:10 ~ 14:00
|
Sequence-level Transcriptome Analysis
- Novel Transcript Discovery
- Alternative Splicing Identification
- RNA-editing Analysis
- New/Fusion Gene Identification
|
|
|
14:10 ~ 15:30
|
실 습 II: Alternative Splicing Identification
RNA Editing Site Annotation
Fusion Gene Identification
|
이수연s, 임재현
|
|
15:40 ~ 16:30
|
Non-coding RNAs in RNA-Seq Data
- miRNA Expression Profiling
- miRNA Target Gene Prediction
- Non-coding RNA Characterization
|
남진우 교수
(한양대)
|
|
16:40 ~ 18:00
|
실 습 III: miRNA Sequencing Data Process
miRNA Expression Profiling
non-coding RNA Resources
|
서희원, 임영균
|
|
DAY 4: Exome Sequencing and Cancer Genome Bioinformatics
|
|
8월 29일(목)
|
|
시간
|
주 제
|
강 사
|
|
8:30 ~ 9:30
|
등록 및 사전 프로그램 설치 |
|
9:30 ~ 9:50
|
Exome Sequencing and Cancer Genome Bioinformatics
|
김주한 교수
|
|
9:50 ~ 10:40
|
Exome Sequencing and Rare Disease
- Exome Sequencing Data
- Exome Sequencing of Rare Disease
- Variant Analysis and Annotation
|
김남신 박사
(생명공학연구원KOBIC)
|
|
10:50 ~ 12:10
|
실 습 I: Trio-Exome-Sequencing Data Analysis
Known Variant Filtering
Detection of Disease-causing Variations
(SIFT, PolyPhen2, VAAST)
Disease Gene Prioritization
|
서희원, 김기태
|
|
12:10 ~ 13:10
|
중 식
|
|
13:10 ~ 14:00
|
Cancer Genome Bioinformatics
- Cancer Genome Analysis
- Identifying Genomic Rearrangement
- Gene Fusion Analysis
- Survival Analysis
|
송영수 교수
(한양의대)
|
|
14:10 ~ 15:30
|
실 습 II: TCGA Data Analysis
Cancer Genome Analysis (Multiple Plotting,
Network Analysis, Visualization, Mutation,
Methylation, Survival Analysis)
Cancer Panel, OncoMap
|
임재현, 고인석
|
|
15:40 ~ 16:30
|
Copy Number and Genomic Rearrangement
- CNA Identification in Cancer Genome
- Copy Number Data Processing
- Genomic Rearrangement
|
김봉조 박사
(국립보건연구원)
|
|
16:40 ~ 18:00
|
실 습 III: Cancer Genomic Rearrangement
Identification of CNV Regions
CNV Database
|
임재현, 임영균
|
|
DAY 5: GWAS and Post-GWAS, eQTL, PheWAS and EWAS Data Analysis
|
|
8월 30일(금)
|
|
시간
|
주 제
|
강 사
|
|
8:30 ~ 9:30
|
등록 및 사전 프로그램 설치 |
|
9:30 ~ 9:50
|
GWAS and Post-GWAS, eQTL, PheWAS and EWAS Data Analysis
|
김주한 교수
|
|
9:50 ~ 10:40
|
SNPs Data Analysis and Genome Wide Association Study
- Linkage Disequilibrium Analysis
- Genotype & Haplotype
- Rare Variant Analysis
- Regression-based Testing
|
이채영 교수
(숭실대)
|
|
10:50 ~ 12:10
|
실 습 I: Haplotype Estimation, LD Blocking
GWAS Catalog
GWAS test with PLINK software
|
임영균, 박찬희
|
|
12:10 ~ 13:10
|
중 식
|
|
13:10 ~ 14:00
|
Post-GWAS and eQTL Data Analysis
- Post-GWAS: Connection to GWAS
- Runs of Homozygosity (ROH)
- Cis- and trans-expression QTL
- eQTL Hotspots
|
박이영 박사
(서울의대)
|
|
14:10 ~ 15:30
|
실 습 II: Idenfity eQTL hotspots
eQTL Resources
|
임재현, 김기태
|
|
15:40 ~ 16:30
|
PheWAS, EWAS and Electronic Medical Records Data Analysis
- EMR and beyond GWAS
- Phenome-Wide Association Study (PheWAS)
- Environment-Wide Association Study (EWAS)
|
김주한 교수
(서울의대)
|
|
16:40 ~ 18:00
|
실 습 III: EMR Data Structure and Extraction
SNPs and Disease Associations from EMR
EMR Based Phe-WAS
|
고인석, 이수현
|
|
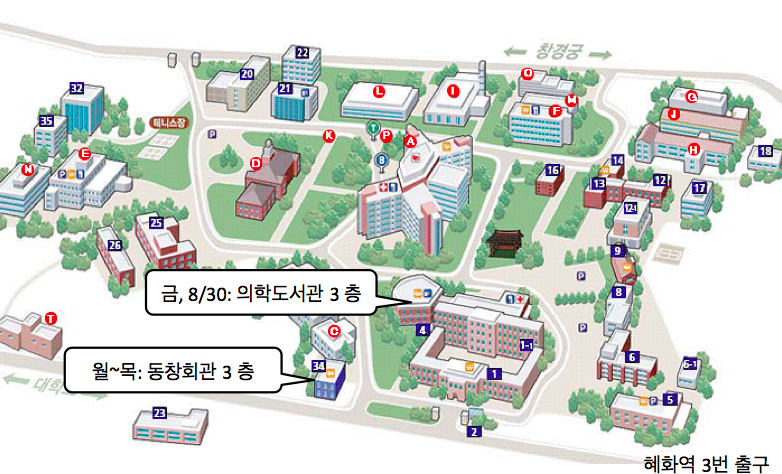
오시는 길
|
교육장소 |
서울의대 동창회관 3층 가천홀/의학도서관 3층, 서울시 종로구 연건동 28
교 통 | 지하철 4호선 혜화역 3번 출구 이용
|
|






